Proposed Structure-Based Model of How Interplay between PTMs Influences Tau Filament Structure. Based on our cryo-EM maps and MS PTM mapping onto atomic models, we conclude that ubiquitination of tau can mediate inter-protofilament interfaces in the doublet CBD fibril and straight filament from AD. If sites on tau favoring the formation of doublet fibrils in CBD (K353) or straight filaments in AD (K311 and K317/321) are acetylated, or ubiquitinated with low occupancy, this inter-protofilament interface is less likely to form. The singlet fibril in CBD does not bind to a second protofilament, and paired helical filaments in AD have structures that do not require mediation by non-tau components at their inter-protofilament interface. The outcome of this model is that the incorporation of ubiquitin into tau filaments in CBD and AD mediates inter-protofilament packing resulting in distinct ultrastructural polymorphs, tuning the ratio of fibril subtypes in tau inclusions.
Structural dynamics by 4D Electron Crystallography / Micro-Electron Diffraction. 4D Images and diffraction patterns of a single crystal are recorded at various tilt-angles using a time-delay tn. Amplitudes from the diffraction patterns are combined with phases from the images and after merging data together from a wide variety of tilt angles, a three-dimensional structure at time-delay tn is generated. This procedure is then repeated for the next time delay, tn+1, by changing the optical path length of the pump beam and recording a tilt series and solving the 3D structure at this later time. The 3D structures solved at multiple time delays can be put together sequentially to form frames of a movie showing the conformational changes of the protein of interest. Image adapted with permission from: Membrane protein structure determination by electron crystallography. I. Ubarretxena-Belandia & D.L. Stokes. Current Opinion in Structural Biology 08/2012; 22(4):520-528.
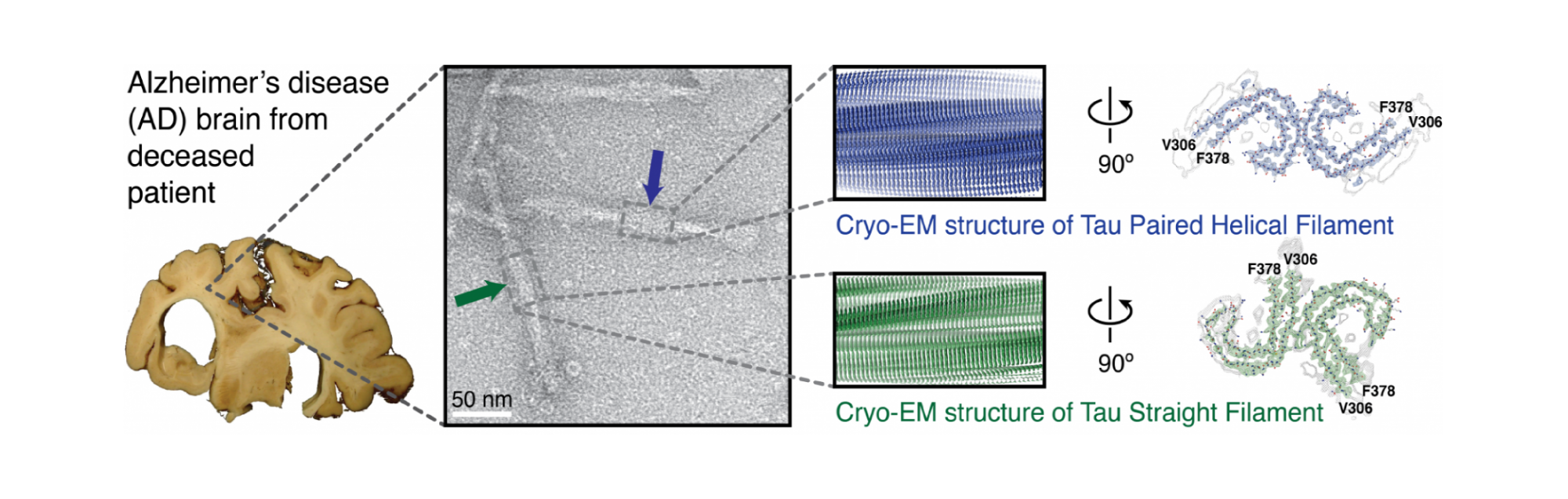
Cross-β structure and tau deposits in Alzheimer’s disease brain. From left to right, the illustration shows the Alzheimer's disease brain used for cryo-EM, an electron micrograph of negatively stained filaments with a blue arrow indicating a paired helical filament (PHF) and a green arrow indicating a straight filament (SF), cryo-EM reconstructions of PHFs (blue) and SFs (green) with detailed cross sections, and de novo atomic models of filaments showing C-shaped subunits stacked to form each protofilament, with protofilaments paired into twisted polymorphic fibrils.
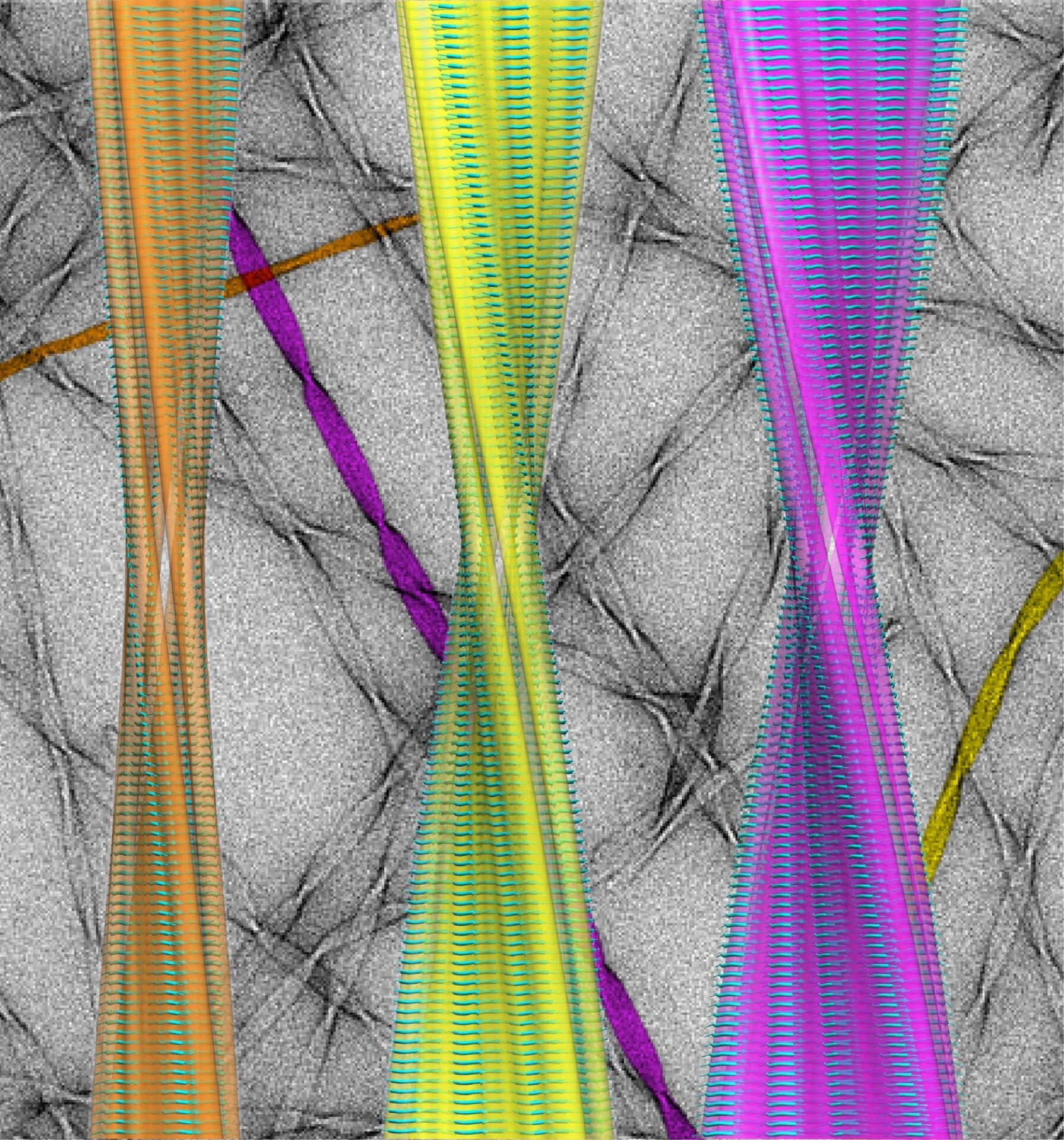
Atomic Structure of a Family of Amyloid Fibrils: pictured are the atomic-resolution structures of three amyloid polymorphs against a (falsely coloured) background image of the fibrils taken with a transmission electron microscope. Determining the fibril structures, and defining the major structural elements and interactions contributing to their hierarchical self-assembly, provides insight into the formation of polymorphic amyloid in a range of protein deposition disorders including Alzheimer's and Parkinson's diseases.
Cover story in Popular Mechanics highlights the work of Dr. Fitzpatrick on Patient-Based Structural Biology of Neurodegenerative Diseases using Cryo-Electron Microscopy.
The image shows a paired helical filament of tau protein, which is the major component of tangles in Alzheimer’s disease. Tau filaments make up the protein inclusions seen inside nerve cells in the brains of people with many neurodegenerative conditions, including Alzheimer’s. The filaments were isolated from the brain of an individual with Alzheimer’s and imaged using cryo-electron microscopy. Individual tau proteins form C shapes (white and blue), which stack together to form filaments. The high-resolution structures of these filaments may help the development of novel diagnostics and therapeutic compounds.
The Fitzpatrick Lab SEM has arrived! Excited for Volume Electron Microscopy 3D reconstructions of Alzheimer's disease, Parkinson's disease and ALS tissue.
Structure of an amyloid fibril at atomic resolution. The structure shown is of one of several polymorphs of amyloid fibrils that are formed in vitro. The combination of cryo‑electron microscopy imaging with solid-state NMR analysis has enabled the determination of an atomic-level structure..
AT8-stained Tau aggregation (green) around DAPI-stained nuclei (blue) at x60 magnification.